Lecture 22: Profiling and microbenchmarking
An order of operations for programming
- Make it run
- Make it right
- Make it fast
When speed matters
- You are working with very large data
- You are running a process (a simulation, a data analysis, etc.) many times
- A piece of code will be called many times (e.g., choosing a split in a decision tree)
Goals
- Learn how to identify bottlenecks in code
- Learn approaches for more efficient code in R
- Time permitting: learn how to use C++ to make code faster
Example: timing code
Suppose we want to compute the mean of each column of a data frame:
Example: timing code
Suppose we want to compute the mean of each column of a data frame:
Alternatives
What are the alternatives to this for-loop approach?
Alternatives
Comparing performance
Microbenchmarking: Evaluating the performance of a small piece of code
bench::mark(
means <- for_loop_means(data),
means <- apply(data, 2, mean),
means <- colMeans(data),
check = F
)# A tibble: 3 × 6
expression min median `itr/sec` mem_alloc `gc/sec`
<bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
1 means <- for_loop_means(data) 1.93s 1.93s 0.519 1.85KB 0
2 means <- apply(data, 2, mean) 2.03s 2.03s 0.493 400.57MB 0.987
3 means <- colMeans(data) 461.4ms 469.34ms 2.13 114.45MB 1.07 Profiling
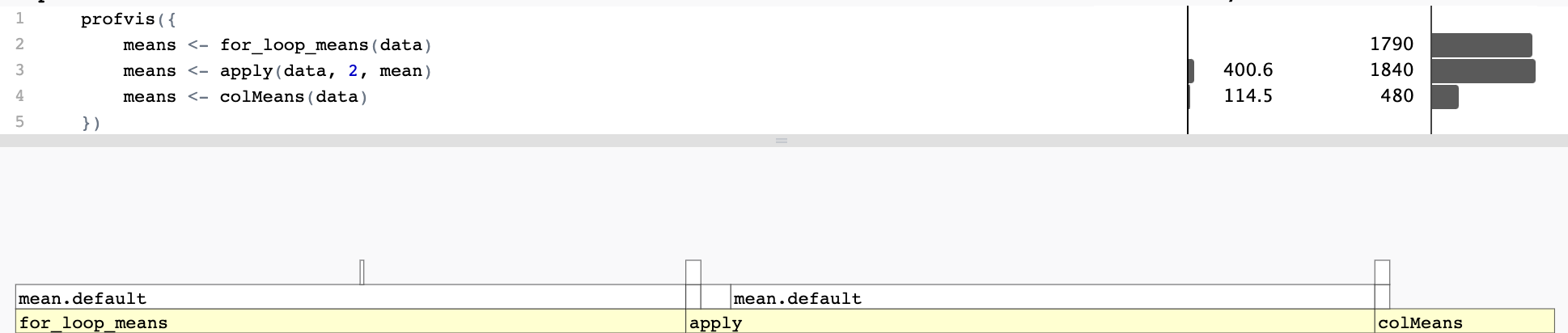
Space for efficiency increases?
function (x, na.rm = FALSE, dims = 1L)
{
if (is.data.frame(x))
x <- as.matrix(x)
if (!is.array(x) || length(dn <- dim(x)) < 2L)
stop("'x' must be an array of at least two dimensions")
if (dims < 1L || dims > length(dn) - 1L)
stop("invalid 'dims'")
n <- prod(dn[id <- seq_len(dims)])
dn <- dn[-id]
z <- if (is.complex(x))
.Internal(colMeans(Re(x), n, prod(dn), na.rm)) + (0+1i) *
.Internal(colMeans(Im(x), n, prod(dn), na.rm))
else .Internal(colMeans(x, n, prod(dn), na.rm))
if (length(dn) > 1L) {
dim(z) <- dn
dimnames(z) <- dimnames(x)[-id]
}
else names(z) <- dimnames(x)[[dims + 1L]]
z
}
<bytecode: 0x7fe4b5412cc0>
<environment: namespace:base>Increase efficiency by avoiding extraneous steps
n <- 100000
cols <- 150
data_mat <- matrix(rnorm(n * cols, mean = 5), ncol = cols)
data <- as.data.frame(data_mat)
bench::mark(
means <- colMeans(data_mat),
means <- colMeans(data),
check = F
)# A tibble: 2 × 6
expression min median `itr/sec` mem_alloc `gc/sec`
<bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
1 means <- colMeans(data_mat) 437ms 437ms 2.29 1.22KB 0
2 means <- colMeans(data) 453ms 453ms 2.21 114.45MB 2.21Profiling
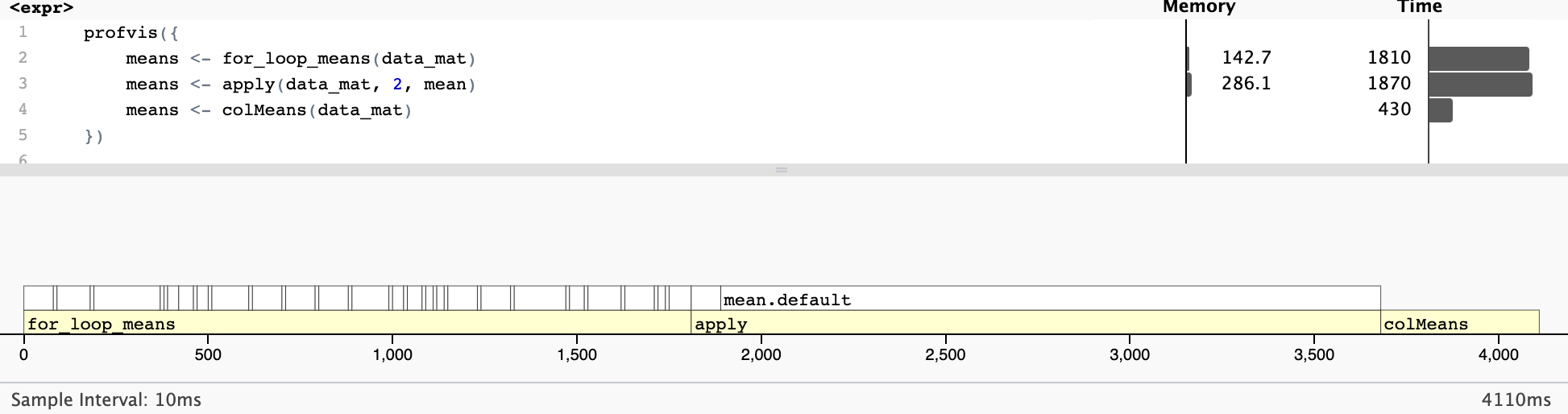
Profiling


Class activity
https://sta279-f23.github.io/class_activities/ca_lecture_24.html