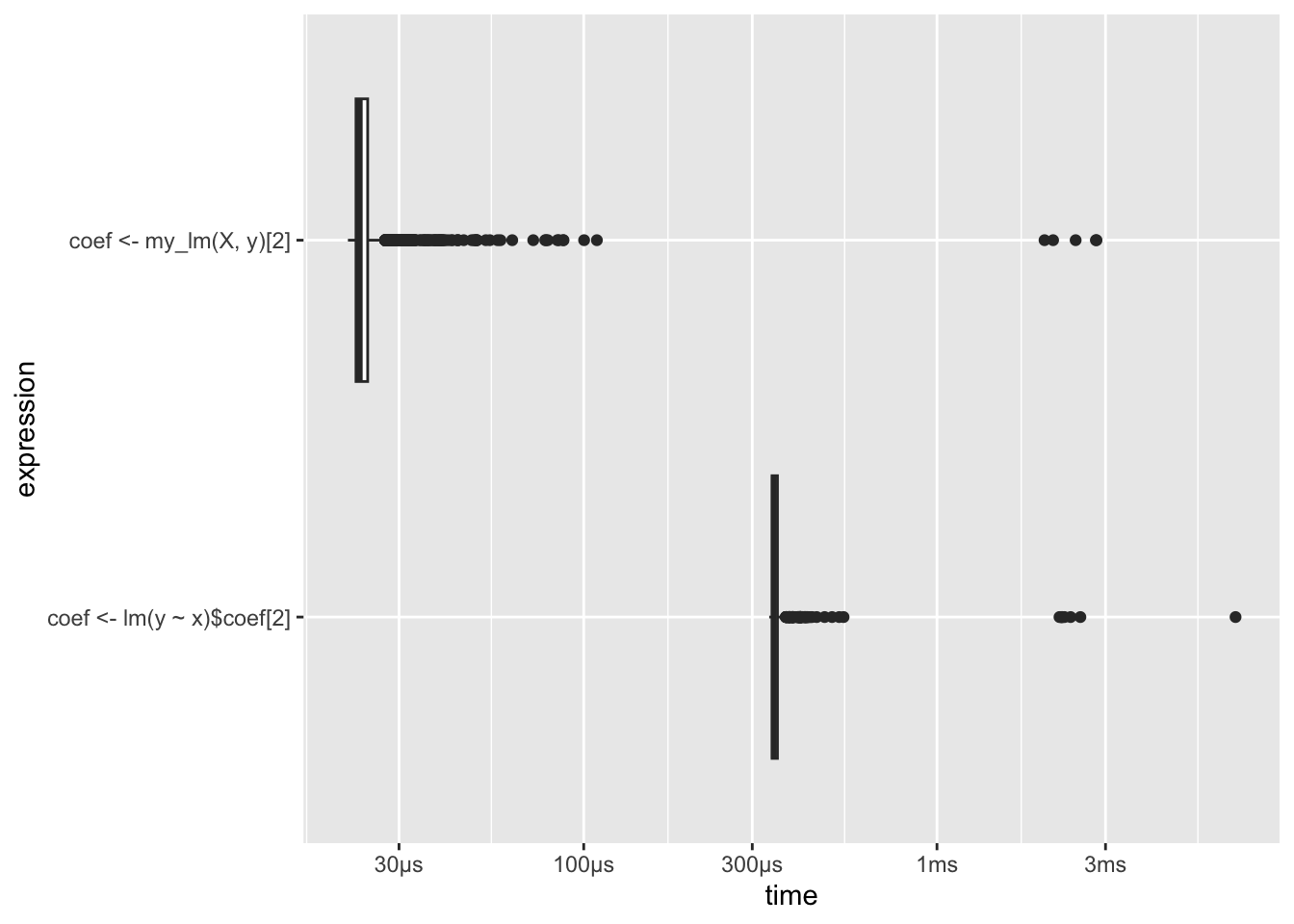
system.time({
nsim <- 500
n <- 100
beta0 <- 1
beta1 <- 0.5
save <- vector()
for (i in 1:nsim) {
x <- rnorm(n)
y <- beta0 + beta1*x + rnorm(n)
coef <- lm(y ~ x)$coef[2]
save <- c(save, coef)
}
save <- data.frame(save)
}) user system elapsed
0.200 0.012 0.211